An international collaboration of scientists identified four fragments that interact with the nsp10 protein of the SARS-CoV-2 virus using the FragMAX platform and BioMAX beamline. The fragments could be used to develop inhibitors that supplant key enzymes activated by the protein—an application which holds potential to block the viral replication process.
Drug development for Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) and future coronaviruses has emerged as a distinct, next-step solution alongside the evolving development of Covid-19 vaccines worldwide. The unknowns of how long protection lasts with vaccination, the appearance of new virus variants, and a lack of existing, effective drugs against Covid-19, have increased the urgency for development of targeted, SARS-CoV-2 medications. MAX IV’s ligand search platform FragMAX is designed to deliver solution-level answers.
“The FragMAX experiment is a screening experiment. We get a direct visualization of the fragment interaction with the protein, with fine atomic details and complexity revealed and rationalized in a single experiment,” said Vladimir O. Talibov, former Beamline Scientist at BioMAX and study author. “X-ray crystallographic fragment screenings are extremely information-rich, and a positive outcome constitutes not only discovery of novel ligands. Obtained data has a very strong impact on further activities and is essential for improvement of the compounds in a potential drug discovery campaign.”
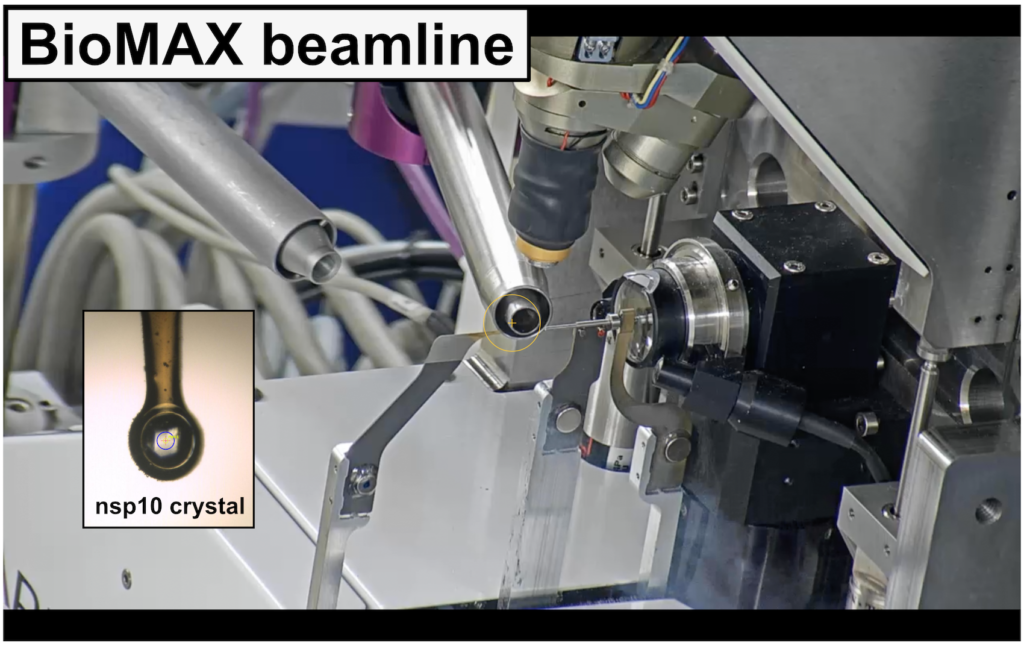
The study draws from earlier work initiated by Professor Frank Kozielski of University College London who, at the start of the Covid-19 pandemic, was a visiting research fellow at Lund University. The research group solved the structure of the nsp10 protein with diffraction data from BioMAX beamline at MAX IV Laboratory.
Finding a match
The current study, published in RSC Chemical Biology, is the result of the concerted efforts of the Lund Protein Production Platform (LP3) at Lund University, Deuteration and Macromolecular Crystallisation (DEMAX) group at ESS and MAX IV’s FragMAX platform and BioMAX beamline team.
The experiment ran from late October to early November 2020, beginning with crystallization of the nsp10 protein by the LP3 facility. The research group including MAX IV scientist Vladimir O. Talibov— who designed and assembled the full FragMAX fragment library—screened the library in search of small molecules that interact with the viral protein. The team at BioMAX assisted with crystal data collection, and complete data analysis was done with the FragMAXapp, a software program developed at MAX IV that runs on the laboratory computer cluster. The DEMAX group supported with protein modelling, and experimental design and analysis.
“FragMAX, DEMAX and LP3 share lab space at Biologihuset and therefore have the entire gene-to-hit workflow under one roof. This arrangement provides an important link between the structural biology community at Lund University and MAX IV and in the future with ESS,” said Tobias Krojer, MAX IV Scientist and FragMAX manager. “It’s vital that the project is collaborative and multi-disciplinary, just the way drug development works. I’m pleased we have established a generic and robust workflow at FragMAX that can be routinely applied to a variety of projects.”
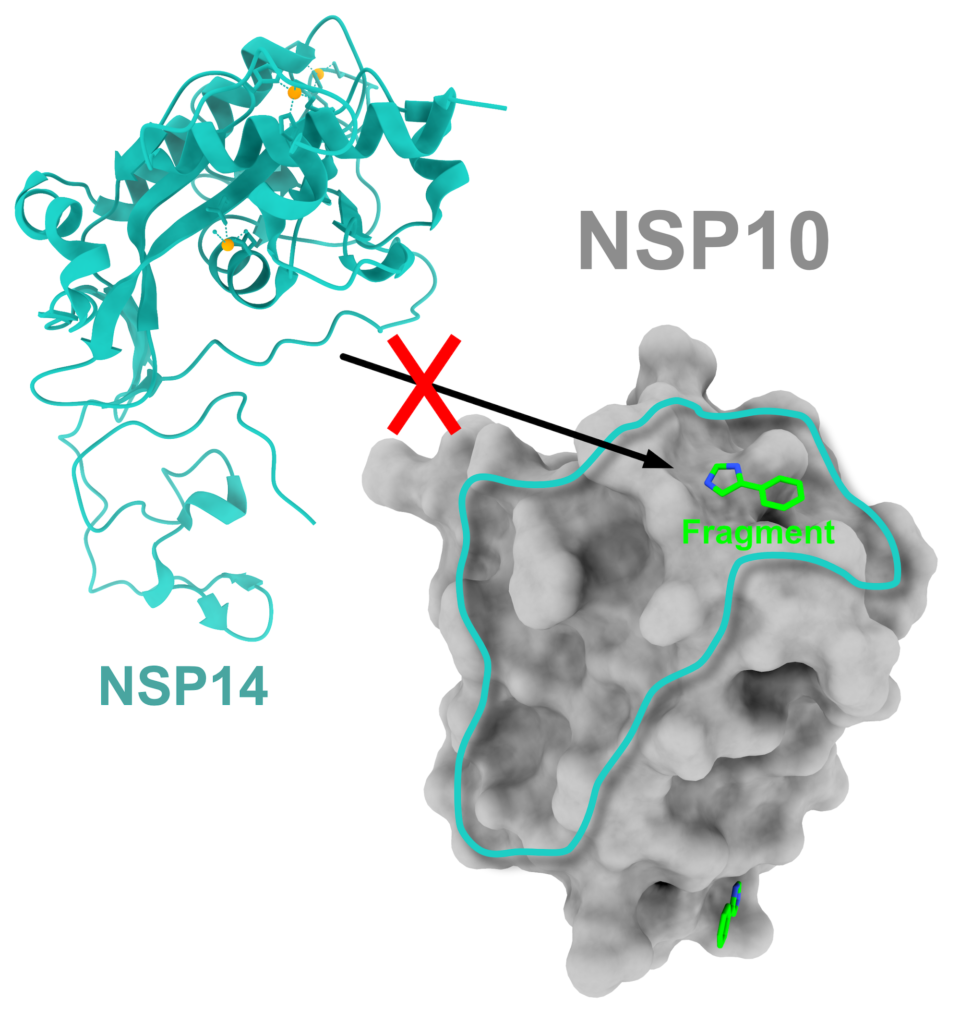
The researchers used nano differential scanning fluorometry, X-ray crystallography and microscale thermophoresis in their analysis. Four fragments were matched to the nsp10 protein, with binding at two specific regions—the nsp14–nsp10 complex interface and the nsp16–nsp10 complex interface. Nsp10 is known to promote the activity of enzymes nsp14 and nsp16, both crucial for viral replication. It is proposed that small molecules similar to the discovered fragments could bind to nsp10, consequently blocking its function and suppressing the virus life cycle. Project data is available for scientists in the global Protein Data Bank.
“The experiment owes a lot to BioMAX beamline, especially to the data collection rates provided by its state-of-the-art equipment, high quality data acquisition and multiple well-synchronized processes that provide a smooth user experience. The results were achievable thanks to the BioMAX team, MAX IV IT team and MAX IV high-performance computing infrastructure,” said Talibov.
Related links:
COVID-19 Data Portal Sweden
Fast Access user portal — SARS-CoV-2 & urgent research