An international collaboration between the UCL School of Pharmacy, the Lund Protein Production Platform (LP3) and ESS, through its DEMAX platform, have performed biophysical and structural studies of three non-structural proteins from the novel coronavirus, SARS CoV-2, the causative agent of COVID-19. In the spring of 2020, they managed to solve and started to analyse one of these proteins, Nsp10, by using the BioMAX beamline at MAX IV Laboratory. Early October published their results in the International Journal of Molecular Sciences.
To be able to find an efficient drug, which prohibits the novel coronavirus from causing the disease COVID-19, one important aim is to understand how to block the virus from replicating its genomic material. By doing so the virus will “die out” over time as it fails to reproduce or make infectious particles. One way of getting this knowledge is to use x-rays to obtain an image of the structure of the proteins involved in this process.

There are 16 non-structural proteins (NSPs) in SARS CoV-2 that play important roles in viral replication and transcription. The virus has excellent proofreading capabilities leading to a low mutation rate in viral replication. Interfering with the virus’s ability to replicate and produce a mature, infectious virus capsid is a key focus for many international researchers. High-resolution crystallographic studies play an important role in finding either new inhibitors or studying how existing drugs can be repurposed to block the novel coronavirus.
There are several proteins involved with proofreading of the viral RNA and its protection by RNA cap methylation, ensuring RNA replication. The research team’s studies focus on three of these: Nsp10, Nsp14, and Nsp16. A key part of this work is to establish crystallization conditions to enable high-throughput fragment-based screening in the search for novel small molecule inhibitors that can block the viral replication cycle.

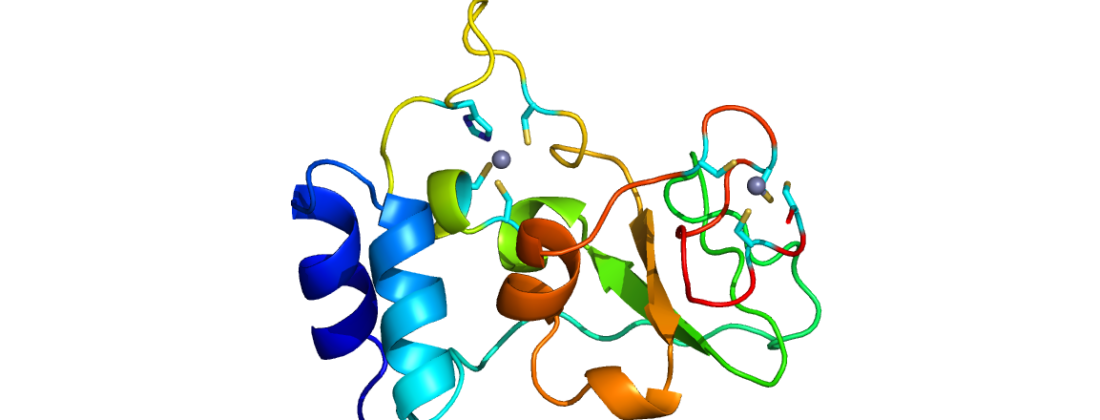
During spring 2020 the research team were able to produce crystals of one of these proteins, Nsp10 (Figure 1a & 1b).
Using rapid access, the data were collected remotely at the BioMAX beamline at MAX IV. The relevant data have been shared with the research community at the Covid-19 data portal Sweden. The deposited data (PDB:6ZCT; Figure 2) can be found listed in the protein data module. By ongoing optimization, the structure was ultimately solved to 1.6 Å (PDB ID:6ZPE).
The ultimate goal for this research project is to obtain high-resolution X-ray crystal structures of these proteins alone or in complex with each other to enable the search for small molecule inhibitors that disrupt their activity. These structural studies will also be complemented with other biophysical characterization experiments such as thermal stability, microscale thermophoresis and solution scattering studies using X-rays and neutrons. The other work on Nsp14 and 16 also continues and the research team are working towards crystal structures of those as well.
This project is an international collaboration between Prof. Frank Kozielski (Main PI, School of Pharmacy, University College London), Dr Wolfgang Knecht (Lund Protein Production Platform (LP3), Lund University) and Dr Zoë Fisher (DEMAX, European Spallation Source ERIC, Lund). The project is supported by the MAX IV rapid access call COVID-19, and the ESS DEMAX call for prioritized projects related to COVID-19 research.
Acknowledgements: We also need to thank all Lund Protein Production Platform (LP3) staff for excellent support and performing all experiments. We thank MAX IV Laboratory for rapid beamtime access at BioMAX through its COVID-19 response actions. We thank the BioMAX beamline staff for excellent support. We acknowledge the support of the project by the ESS for projects related to COVID-19 as well as to Lund University and its faculties for support to LP3.