Fungal enzymes play an important role in the breakdown of plant cell walls during plant degradation. An international collaboration of researchers explored the auxiliary activities 7 (AA7) enzyme family, characterizing four fungal enzymes and uncovering a novel class of flavo-enzymes, exemplified by oligosaccharide dehydrogenase. The enzymes fuel the activity of lytic polysaccharide monooxygenases (LPMOs) in the challenging process of crystalline cellulose degradation. The study, published in Nature Communications, offers promise for tuning the efficiency of enzymatic breakdown processes of biomass feedstocks used in energy and biomaterial production.
“Our work mapped the taxonomic distribution of protein sequences in the AA7 family and revealed a striking high proportion of sequences encoded by major plant pathogenic fungi and Oomycetes,” said Maher Abou Hachem, Professor of Biotechnology and Biomedicine at the Technical University of Denmark (DTU). “Further, the discovered dehydrogenases represent the simplest redox partner that are able to fuel LPMO activity in the absence of exogenous reductants.”
The Abou Hachem lab worked closely with the lab of Research Director Jean-Guy Berrin at the INRAE research institute, Aix-Marseille University and several other research groups
Inspiration for the study came from earlier work by Abou Hachem’s group that analysed a network of oxidative enzymes, including orthologues of AA7 flavo-enzymes and LPMOs, which are co-secreted during fungal growth on polysaccharides.
An evolutionary puzzle
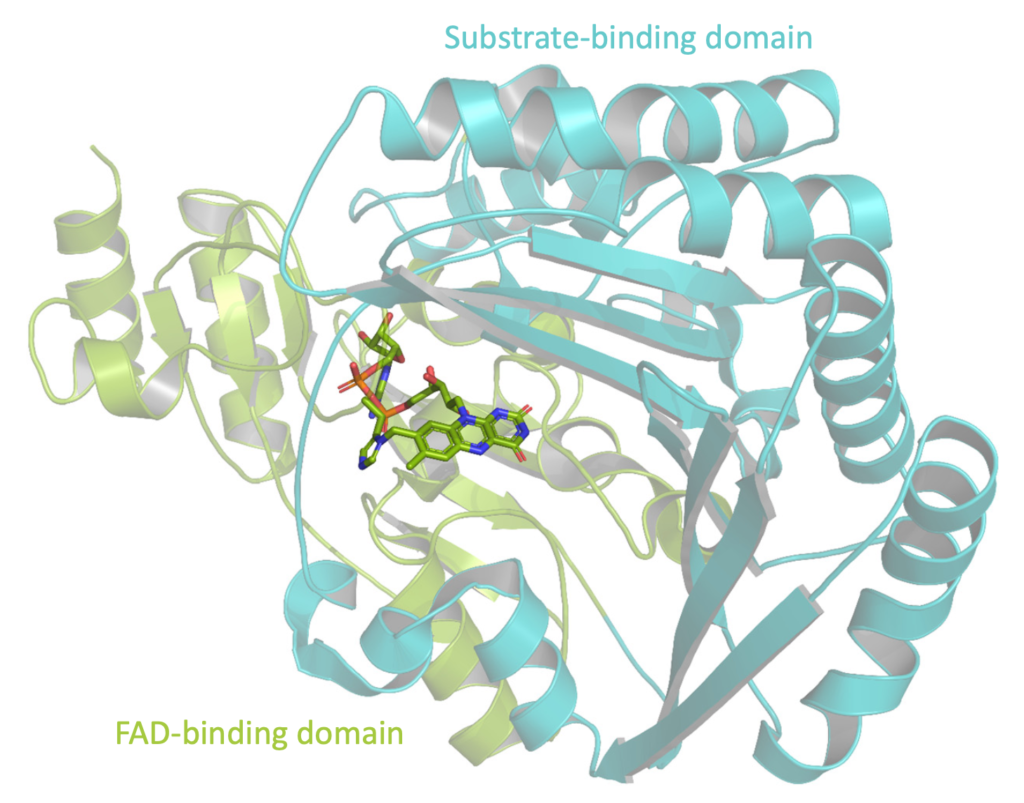
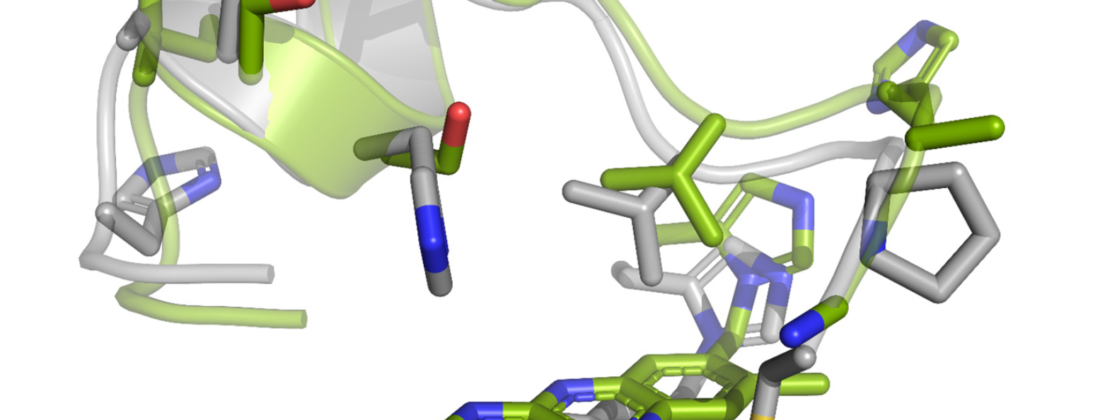
To investigate the genetic diversity of AA7, the research team made an extensive phylogenetic analysis of the fungal family, choosing five enzymes from six clades or genetic groupings for further biochemical and structural study.
An unexpected revelation was that many AA7 gene sequences are encoded by phytopathogenic fungi and lower eukaryotes. Previous research has shown that oligosaccharide oxidases (enzymes) in the AA7 family play a role in plant immunity and resistance to pathogens. Structural data for two enzymes from the fungal pathogen Fusarium graminearum (FgCelDH7C and FgChi7B) was gathered at BioMAX beamline at MAX IV in Lund.
Recent, related studies have used fungal cellobiose dehydrogenase (CDH) as the primary model for priming LPMO enzymes, and hydrogen peroxide as a preferred substrate.
“Based on our previous work and these recent insights into LPMO mechanism, we hypothesized that members of AA7 may serve as redox partners of LPMOs and we went on to test this hypothesis,” said Abou Hachem. The new findings expand the known range of AA7 redox-profiles and viable substrates.
What will it take to design biomass stocks for the future? According to the researchers, solutions lie in greater molecular-level knowledge of how the new-found AA7 dehydrogenases trigger activation of LPMOs, and through engineering of AA7 enzymes with diverse binding and redox capacities.
At present, the lab group has begun new investigations on properties of new AA7 dehydrogenases and seek to gain insight into the interactions between AA7 and LPMOs.