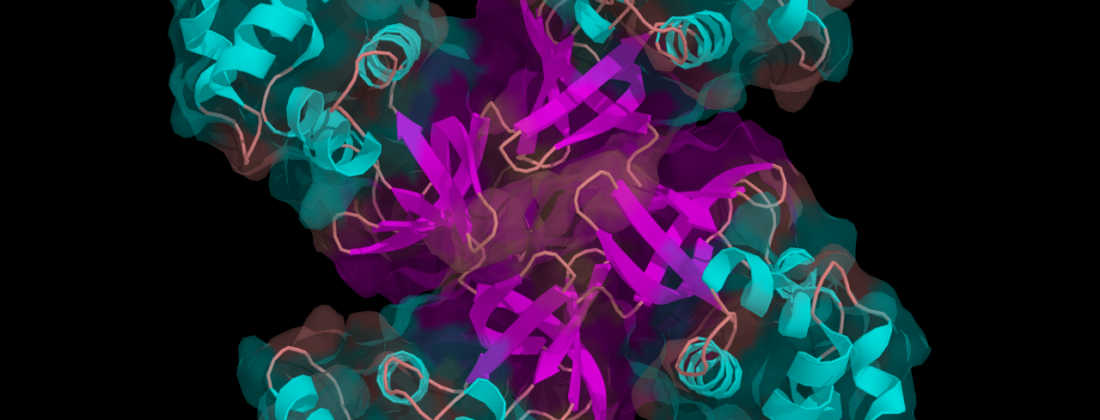
A group of researchers have successfully soaked proteins into large protein crystals, marking a hitherto never reported achievement. The X-ray diffraction data, which were collected on MAX IV’s BioMAX beamline, indicate that the guest proteins could follow at least some of the hosts’ structures signifying a so far unparalleled step towards using crystallographic methods to solve guest protein structures. Such insights could pave the way for major advances in biotechnology, material science, and structural studies.
Soaking smaller molecules into solvent channels of protein crystals has been a widely employed method for obtaining crystalline complexes with ligands, that is ions or molecules that bind to biomolecules. Yet, until now, researchers had fallen short of incorporating guest protein macromolecules and thus revelations about their structure, penetration depth, or distribution.
The data, obtained from the crystals tested via X-ray diffraction on the BioMAX beamline and the observation of guest protein migration via confocal fluorescence microscopy, changes that.
“It was crucial to study many different crystals, so we needed a stable, high-quality beam. The throughput of the measurements from the BioMAX beamline were really fast and the data of really good quality,” said Janina Sprenger, Research Fellow at the Center for Free-Electron Laser Science and lead author of the study which was carried out by a team of scientists from the University of Copenhagen, Lund University, Princeton University, Rutgers University, and the German Electron Synchrotron DESY.
They chose crystals of an Escherichia coli dimeric tryptophan repressor protein (ds-TrpR) as the host protein due to its stable, domain-swapped structure, and monitored the migration of cytochrome c and calmodulin, the latter of which had been tagged fluorescently. After two weeks, they observed that the ds-TrpR’s fluid contents had allowed the two guest proteins to enter its crystal through its solvent channels.
The challenge remaining is that the data does not yet reveal enough protein contents in the right positions to properly investigate their structure. In crystallography, symmetry is key to obtain structural information from a protein. “The host is very symmetric, but we can only solve the structure of the guest if it follows the host’s symmetry and if on almost each symmetric position there is also a guest protein,” Sprenger explained.
In addition, the team is now looking into bioengineering the host crystal to have ‘attachment points’ for the guests in their solvent channels. “It’s like a bait for a guest to attach there,” said Sprenger. “Attachment points go to the right structure and those attachment points follow the symmetry. We hook it to the right position.”
When the scientists examined the ds-TrpR host through X-ray diffraction, they found high concentrations of guest proteins within the ds-TrpR crystal structure after two weeks and noted that the diffraction could reach up to about 2.5 Å. “This resolution is good enough to see biologically relevant details at a more or less atomic level, which means we can see medically relevant molecules that can alter protein functions such as ligands. These are features we see at this range that are very relevant for us,” explained Sprenger.
Even without unlocking the symmetry puzzle, host proteins may function as nano-sized controlled vehicles for the direct delivery of genetic materials, medical drugs, or DNA and RNA carriers to sites within the body. “We know how to modify proteins; we understand better and better how they react with other proteins and molecules. They are natural and – by definition – what life itself takes and uses,” said Sprenger. “We see protein crystals less as a material, but it could be.”
The study report echoed this sentiment stating that protein crystals could even be used as “micro-bioreactors.” In this case, the hosts’ solvent channels make up spaces which would be filled with materials other than guest proteins. Research for such efforts is already under way and could even open the door to the development of materials that do not yet exist. In Sprenger’s words: “Even if you cannot solve the structures from it, there might still be an application for material science as the crystal channels can be explored to be filled with other molecules.”
Publication
Sprenger, J., Carey, J., Schulz, A. et al. Guest-protein incorporation into solvent channels of a protein host crystal (hostal), Acta Cryst. D77, 471-485. (2021), DOI: 10.1107/S2059798321001078
Image Attribution: Phoebus87 at English Wikipedia, CC BY-SA 3.0, via Wikimedia Commons